Contacts
Institute of Science Tokyo
Department of Life Science and Technology
4259-J2-16 Nagatsuta-cho, Midori-ku, Yokohama, 226-8501, JAPAN
Tel: +81-45-924-5136
FAX: +81-45-924-5144
Kohji Seio
Professor
e-mail: kseioatbio.titech.ac.jp
Links
Tokyo Institute of Technology
Department of Life Science
Tokyo Tech Portal
TSUBAME2.5 Portal
IASO R6
Tokyo Tech Electronic Journals and Ebooks List
---
Protein Data Bank
Nucleic Acid Database Server
NCBI PubMed
Web of Knowledge
SciFinder
Google Scholar
Institute of Science Tokyo
Department of Life Science and Technology
4259-J2-16 Nagatsuta-cho, Midori-ku, Yokohama, 226-8501, JAPAN
Tel: +81-45-924-5136
FAX: +81-45-924-5144
Kohji Seio
Professor
e-mail: kseioatbio.titech.ac.jp
Links
Tokyo Institute of Technology
Department of Life Science
Tokyo Tech Portal
TSUBAME2.5 Portal
IASO R6
Tokyo Tech Electronic Journals and Ebooks List
---
Protein Data Bank
Nucleic Acid Database Server
NCBI PubMed
Web of Knowledge
SciFinder
Google Scholar
Recent publications
2025
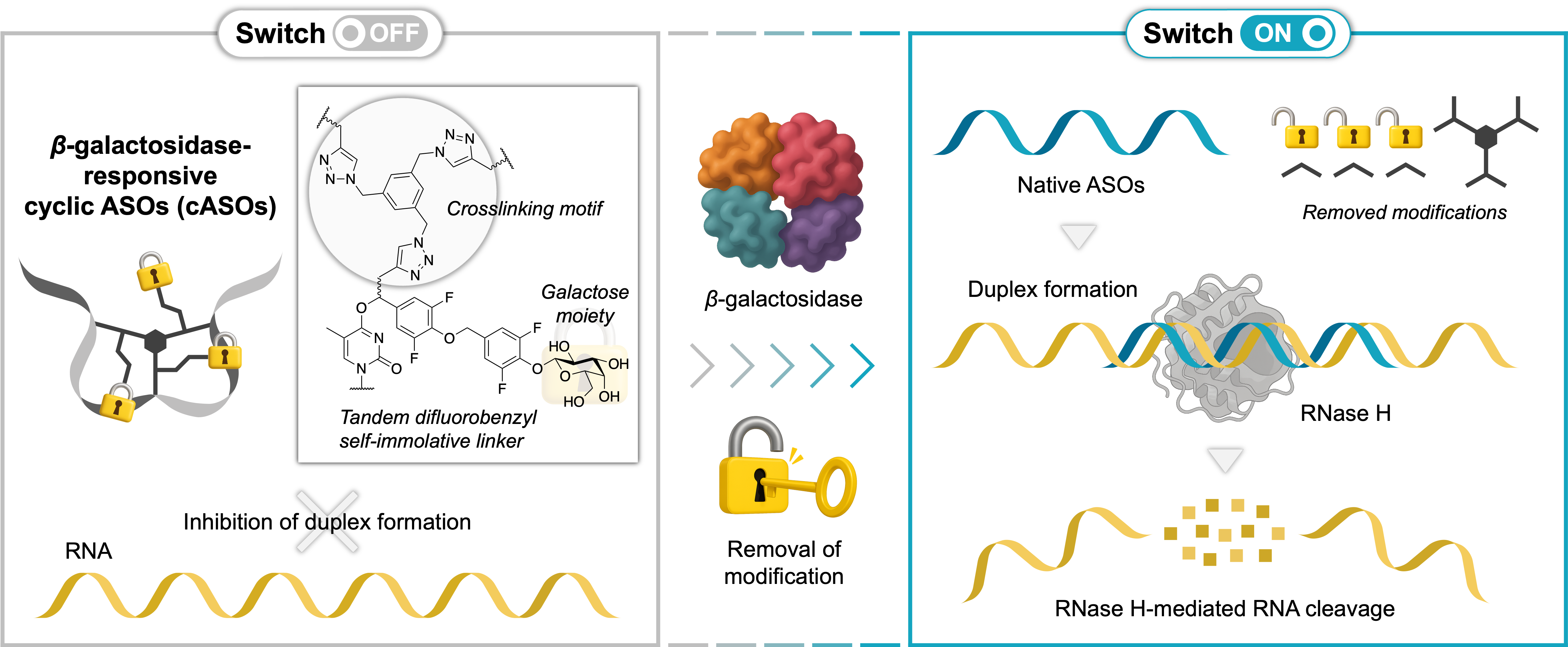
Topological Switch-OFF and β-Galactosidase-Triggered Switch-ON of Cyclic Antisense Oligonucleotides via CuAAC for Controlled RNA Cleavage
Miyaji K, Takeuchi K, Seio K.
Bioconjugate Chem. 2025 Aug 20; 36(8):1820-1837 doi.org/10.1021/acs.bioconjchem.5c00295.
Synthesis of Prodrug-Type Oligonucleotides Modified With a Galactosylated Self-Immolative Linker Cleavable by β-Galactosidase
Miyaji K, Masaki Y, Seio K.
Curr Protoc. 2025 Apr 7;5(4):e70128. doi: 10.1002/cpz1.70128.
Synthesis of LNA gapmers that replace a phosphorothioate linkage with a sulfonamide in the gap region, and their ability to form duplexes with complementary RNA targets
Seio K, Ohnishi R, Tachibana S, Mikagi H, Masaki Y.
Org. Biomol. Chem. 2025 Jan 2;23(2):400-409. doi.org/10.1039/d4ob01350f.
2024
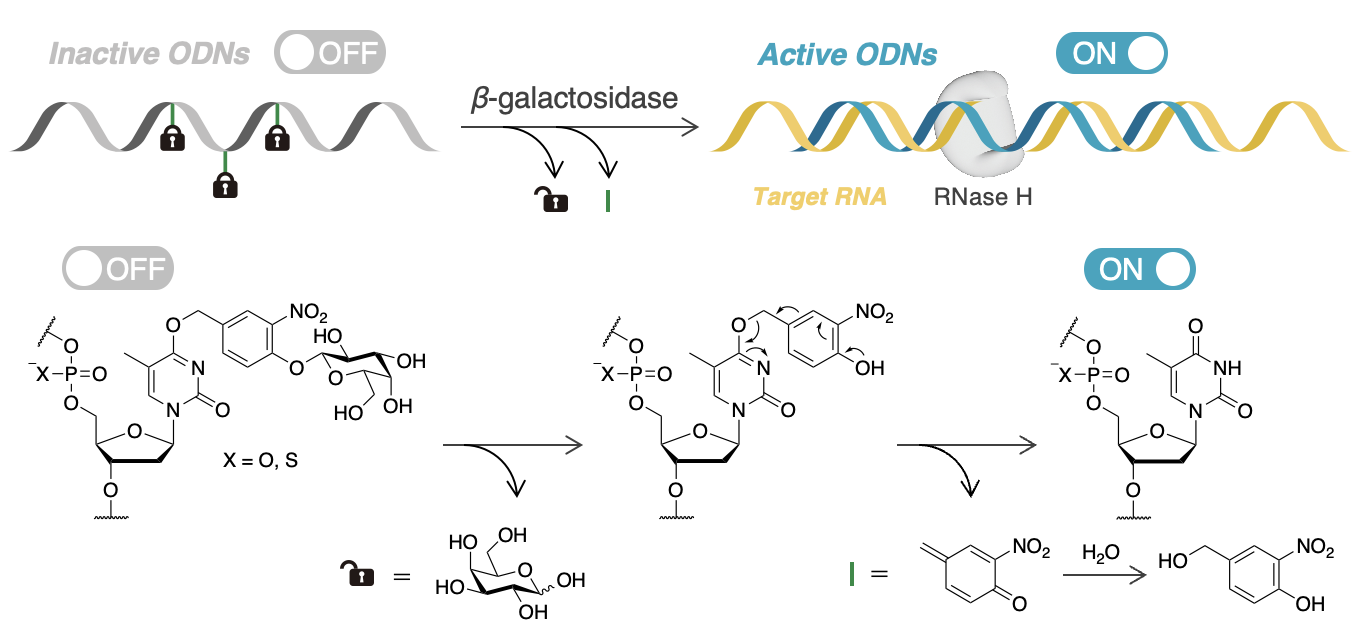
Inhibitory Effects on RNA Binding and RNase H Induction Activity of Prodrug-Type Oligodeoxynucleotides Modified with a Galactosylated Self-Immolative Linker Cleavable by β-Galactosidase
Miyaji K, Masaki Y, Seio K.
Bioconjugate Chem. 2024 Oct 8; 35(10):1587-1607. doi.org/10.1021/acs.bioconjchem.4c00376.
2023
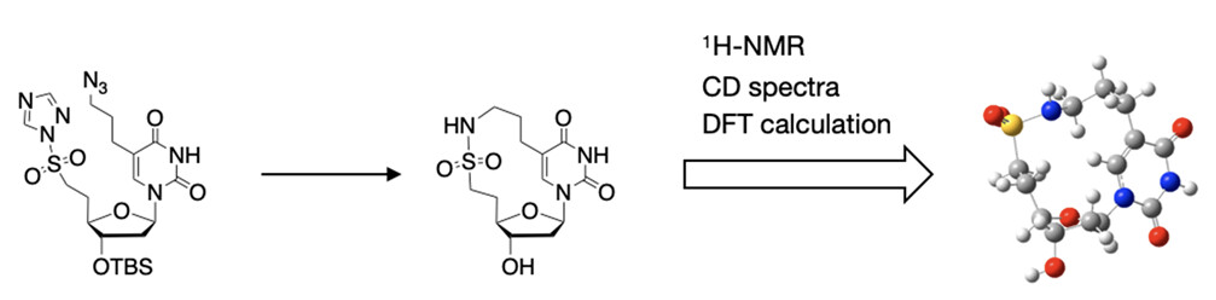
Synthesis and Conformational Analyses of Cyclonucleoside Having 13-Membered Ring Bridging Nucleobase and 5′-Position via a Linker Containing Sulfonamide
Kanagawa T, Tachibana S, Masaki Y, Seio K.
Org. Lett. 2023 Nov 3; 25:7868-7872. doi.org/10.1021/acs.orglett.3c03094.
Alteration of target cleavage patterns and off-target reduction of antisense oligonucleotides incorporating 2-N-carbamoyl- or (2-pyridyl)guanine
Kanagawa T, Koyama A, Masaki Y, Seio K.
Org. Biomol. Chem. 2023 Jun 28; 21:5214-5224. doi.org/10.1039/d3ob00574g.
2022
Insertion of a methylene group into the backbone of an antisense oligonucleotide reveals the importance of deoxyribose recognition by RNase H.
Masaki Y, Tabira A, Hattori S, Wakatsuki S, Seio K.
Org. Biomol Chem. 2022 Oct 27;20:8917-8924. doi.org/10.1039/D2OB01667B.
Quantification of synthetic errors during chemical synthesis of DNA and its suppression by non-canonical nucleosides.
Masaki Y, Onishi Y, Seio K.
Sci Rep. 2022 Jul 15;12(1):12095. doi: 10.1038/s41598-022-16222-2.
Selective and stable base pairing by alkynylated nucleosides featuring a spatially-separated recognition interface.
Okamura H, Trinh G, Dong Z, Masaki Y, Seio K, Nagatsugi F.
Nucleic Acids Res. 2022 Feb 15. doi: 10.1093/nar/gkac140
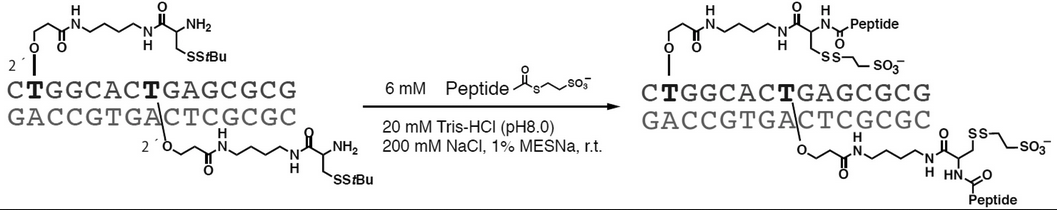
Oligodeoxynucleotides Modified with 2'- O-(Cysteinylaminobutyl)carbamoylethylribothymidine Residues for Native Chemical Ligation with Peptide at Internal Positions.
Masaki Y, Maruyama A, Yoshida K, Tomori T, Kishimura T, Seio K.
Bioconjug. Chem. 2022 Feb 7;33(2):272-278. doi: 10.1021/acs.bioconjchem.1c00575
2021
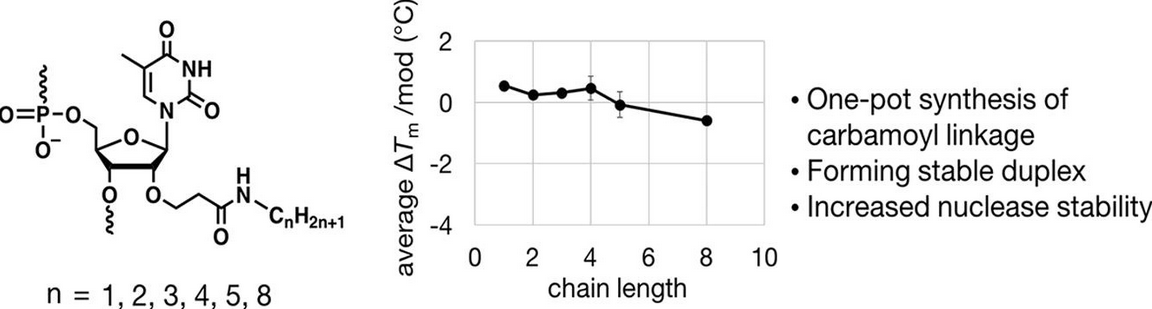
Synthesis of 2′-O-alkylcarbamoylethyl-modified oligonucleotides with enhanced nuclease resistance that form isostable duplexes with complementary RNA.
Kishimura T, Tomori T, Masaki Y, Seio K.
Bioorg Med Chem Lett. 2021 Mar 1;35:127779. doi: 10.1016/j.bmcl.2021.127779
2020
Novel EGFP reporter cell and mouse models for sensitive imaging and quantification of exon skipping
Hara Y, Mizobe Y, Inoue Y, Hashimoto Y, Motohashi N, Masaki Y, Seio K, Takeda S, Nagata T, Wood M, Inoue T, Aoki Y.
Scientific Reports 2020 Jun 22;10. doi: 10.1038/s41598-020-67077-4
Transcription of DNA duplex containing deoxypseudouridine and deoxypseudoisocytidine, and inhibition of transcription by triplex forming oligonucleotide that recognizes the modified duplex
Seio K, Yamaguchi K, Yamazaki A, Kanamori T, Masaki Y.
Nucleosides, Nucleotides & Nucleic Acids 2020 Mar 4;39(6):892-904. doi: 10.1080/15257770.2020.1714652
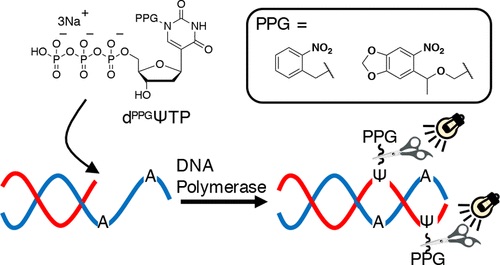
Synthesis of Deoxypseudouridine 5′-Triphosphate Bearing the Photoremovable Protecting Group at the N1 Position Capable of Enzymatic Incorporation to DNA
Takeshita L, Yamada Y, Masaki Y, Seio K.
J. Org. Chem. 2020 Jan 7;85:1861-1870. doi: 10.1021/acs.joc.9b02194
2019
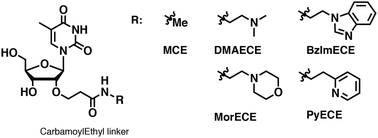
Modification of oligonucleotides with weak basic residues via the 2′-O-carbamoylethyl linker for improving nuclease resistance without loss of duplex stability and antisense activity
Masaki Y, Yamamoto K, Yoshida K, Maruyama A, Tomori T, Iriyama Y, Nakajima H, Kanaki T, Seio K.
Org. Biomol Chem. 2019 Apr 29;17:4835-4842. doi: 10.1039/c9ob00668k
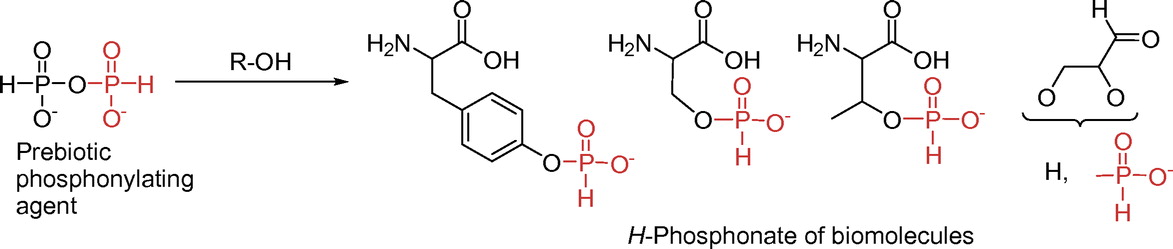
31P NMR Study on the Reactions of Amino Acids and Sugar Derivatives with Pyrophosphorous Acid as a Possible Prebiotic Phosphonylating Agent
Seio K, Shiozawa T, Sugiyama D, Ohno K, Tomori T, Masaki Y.
Bull. Chem. Soc. Jpn. 2019 Apr 10;92(4):905-911. doi: 10.1246/bcsj.20180392
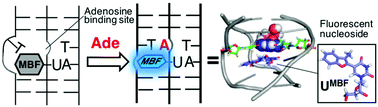
DNA triplex-based fluorescence turn-on sensors for adenosine using a fluorescent molecular rotor 5-(3-methylbenzofuran-2-yl) deoxyuridine
Kanamori T, Masaki Y, Oda Y, Ohzeki H, Ohkubo A, Sekine M, Seio K.
Org. Biomol Chem. 2019 Jan 25;17:2077-2080. doi: 10.1039/C8OB02747A
Synthesis of 2′-O-(N-methylcarbamoylethyl) 5-methyl-2-thiouridine and its application to splice-switching oligonucleotides.
Masaki Y, Yamamoto K, Inde T, Yoshida K, Maruyama A, Nagata T, Tanihata J, Takeda S, Sekine M, Seio K.
Bioorg Med Chem Lett. 2019 Jan 15;29(2):160-163. doi: 10.1016/j.bmcl.2018.12.005
Tolerance of N2-heteroaryl modifications on guanine bases in a DNA G-quadruplex
Masaki Y, Inde T, Maruyama A, Seio K.
Org. Biomol Chem. 2019 Jan 4;17:859-866. doi: 10.1039/C8OB03100B
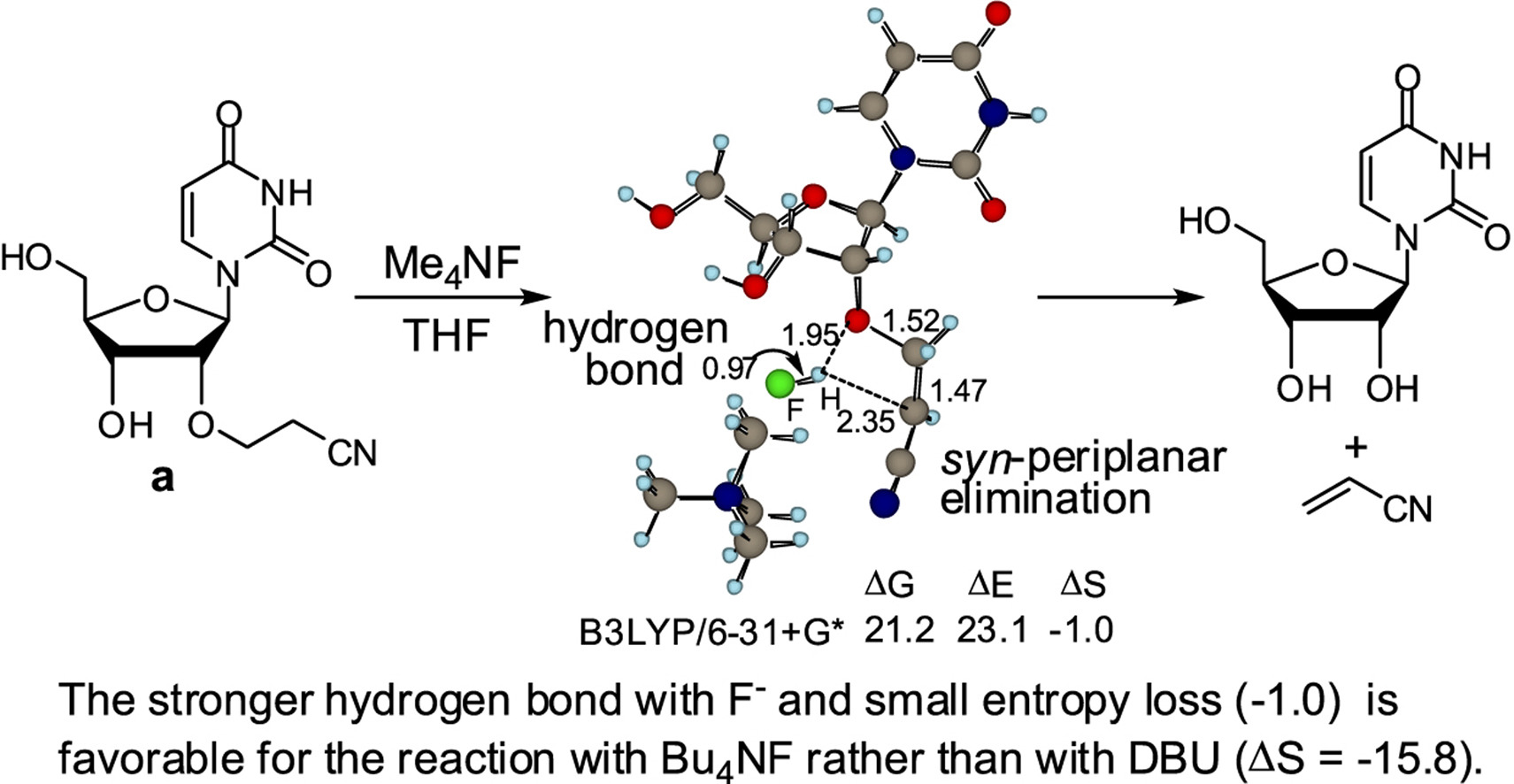
A theoretical study on the elimination reaction of acrylonitrile from 2′-O-cyanoethylated nucleosides by Bu4NF.
Ando K, Saneyoshi H, Seio K, Sekine M.
Tetrahedron 2019 Jan 3;75(1):1-9. doi: 10.1016/j.tet.2018.11.042
2018
Synthesis of and triplex formation in oligonucleotides containing 2'-deoxy-6-thioxanthosine.
Inde T, Nishizawa S, Hattori Y, Kanamori T, Yuasa H, Seio K, Sekine M, Ohkubo A.
Bioorg Med Chem. 2018 Jul 30;26(13):3785-3790. doi: 10.1016/j.bmc.2018.06.004
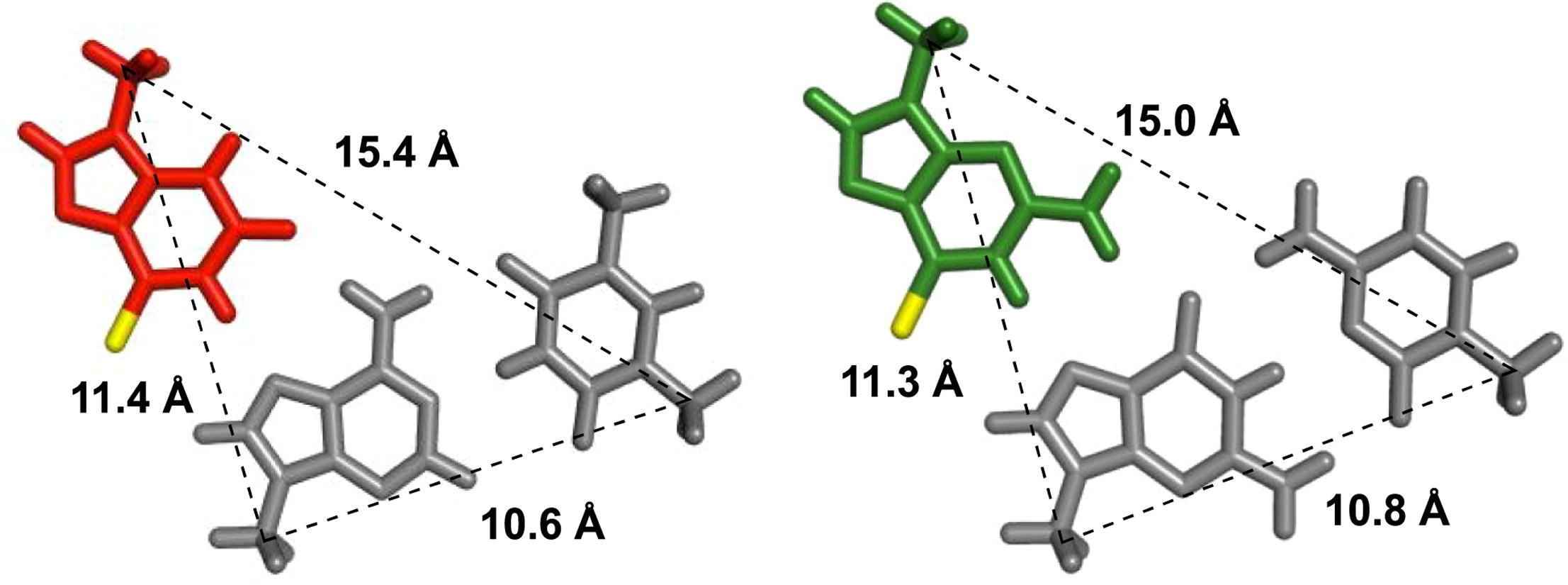
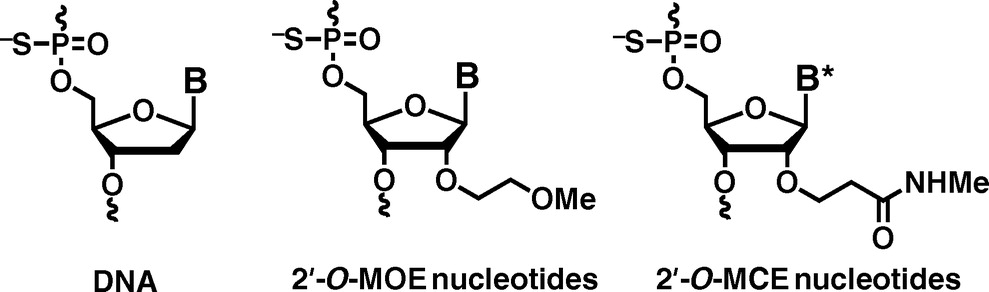
Application of 2'-O-(2-N-Methylcarbamoylethyl) Nucleotides in RNase H-Dependent Antisense Oligonucleotides.
Masaki Y, Iriyama Y, Nakajima H, Kuroda Y, Kanaki T, Furukawa S, Sekine M, Seio K.
Nucleic Acid Ther. 2018 Jul 18;28(5):307-311. doi: 10.1089/nat.2018.0738
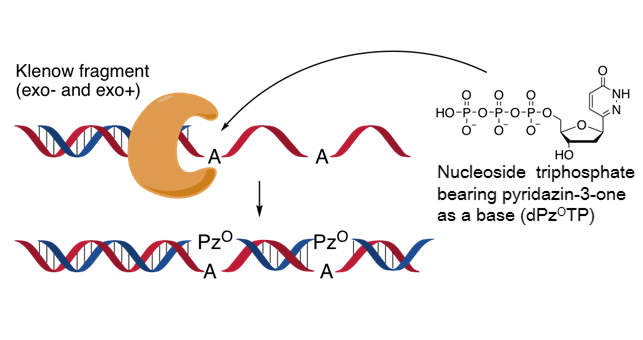
Deoxynucleoside Triphosphate Containing Pyridazin-3-one Aglycon as a Thymidine Triphosphate Substitute for Primer Extension and Chain Elongation by Klenow fragments.
Tomori T, Nagaoka K, Takeshita L, Shiozawa T, Miyatake Y, Masaki Y, Sekine M, Seio K.
J Org Chem. 2018 Jun 28;83(15):8353-8363. doi: 10.1021/acs.joc.8b00918
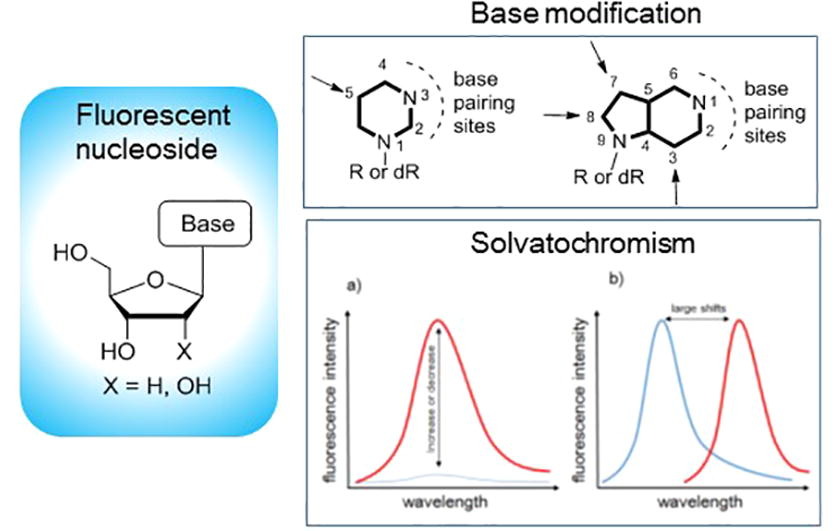
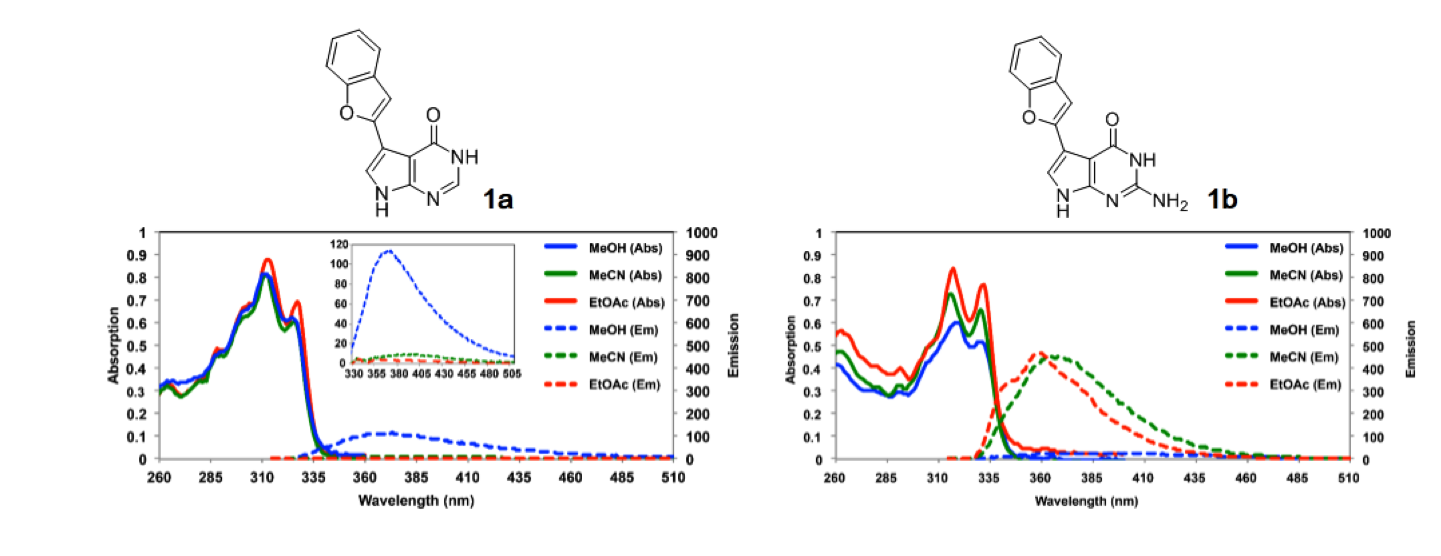
Solvent- and environment-dependent fluorescence of modified nucleobases
Seio K, Kanamori T, Masaki Y.
Tetrahedron Lett. 2018 May 23;59(21):1977-1985. doi: 10.1016/j.tetlet.2018.04.003
2017
Synthesis of photocaged 6-O-(2-nitrobenzyl)guanosine and 4-O-(2-nitrobenzyl) uridine triphosphates for photocontrol of the RNA transcription reaction.
Ohno K, Sugiyama D, Takeshita L, Kanamori T, Masaki Y, Sekine M, Seio K.
Bioorg Med Chem. 2017 Sep 21. pii: S0968-0896(17)31538-9. doi: 10.1016/j.bmc.2017.09.032.
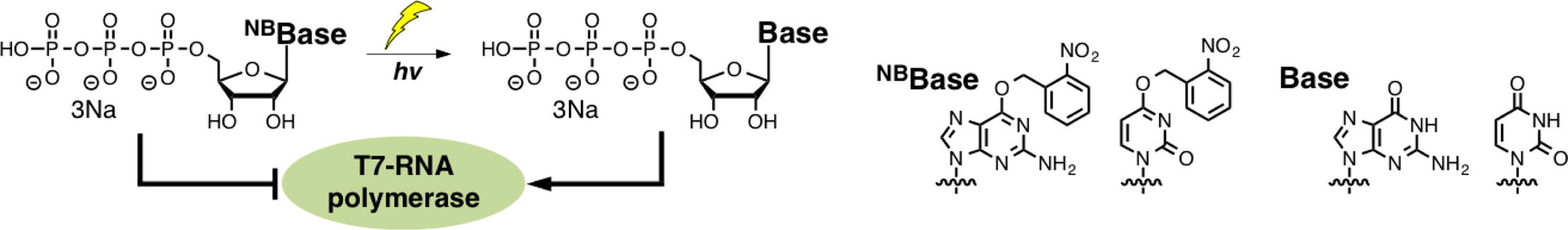
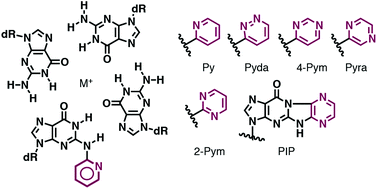
Synthesis of oligonucleotides containing 2-N-heteroarylguanine residues and their effect on duplex/triplex stability.
Inde T, Masaki Y, Maruyama A, Ito Y, Makio N, Miyatake Y, Tomori T, Sekine M, Seio K.
Org Biomol Chem. 2017 Oct 11;15(39):8371-8383. doi: 10.1039/c7ob01875d.
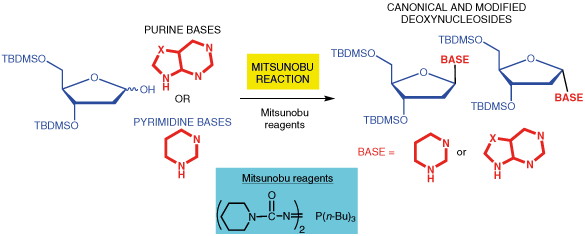
A Systematic Study of the Synthesis of 2ʹ-Deoxynucleosides by Mitsunobu Reaction.
Seio K, Tokugawa M, Kaneko K, Shiozawa T, Masaki, Y.
Synlett 2017; 28(15): 2014-2017. doi: 10.1055/s-0036-1588445.
Deformability Calculation for Estimation of the Relative Stability of Chemically Modified RNA Duplexes.
Masaki Y, Sekine M, Seio K.
Curr Protoc Nucleic Acid Chem. 2017 Mar 2;68:7.27.1-7.27.10.
doi:10.1002/cpnc.25.
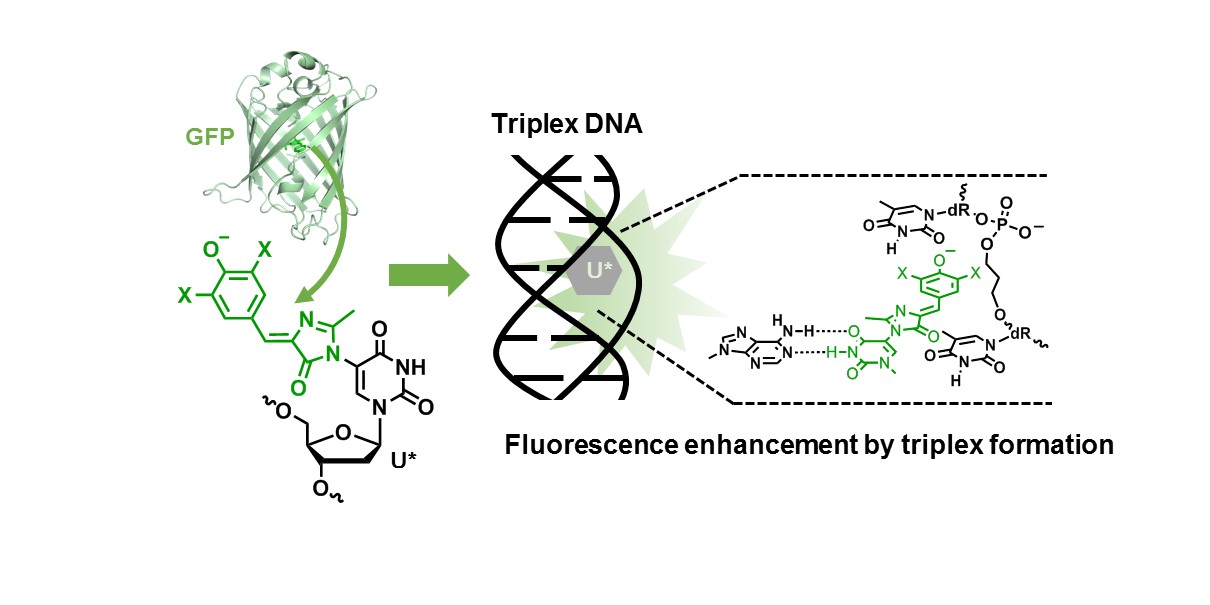
Fluorescence enhancement of oligodeoxynucleotides modified with green fluorescent protein chromophore mimics upon triplex formation.
Kanamori T, Takamura A, Tago N, Masaki Y, Ohkubo A, Sekine M, Seio K.
Org Biomol Chem. 2017 Feb 1;15(5):1190-1197. doi:10.1039/c6ob01278g.
2016
Enzymatic synthesis and reverse transcription of RNAs incorporating 2′-O-carbamoyl uridine triphosphate
Masaki Y, Ito H, Oda Y, Yamazaki K, Tago N, Ohno K, Ishii N, Tsunoda H, Kanamori T, Ohkubo A, Sekine M, Seio K.
Chem Commun (Camb). 2016 Oct 25;52(87):12889-12892. doi:10.1039/C6CC05796A
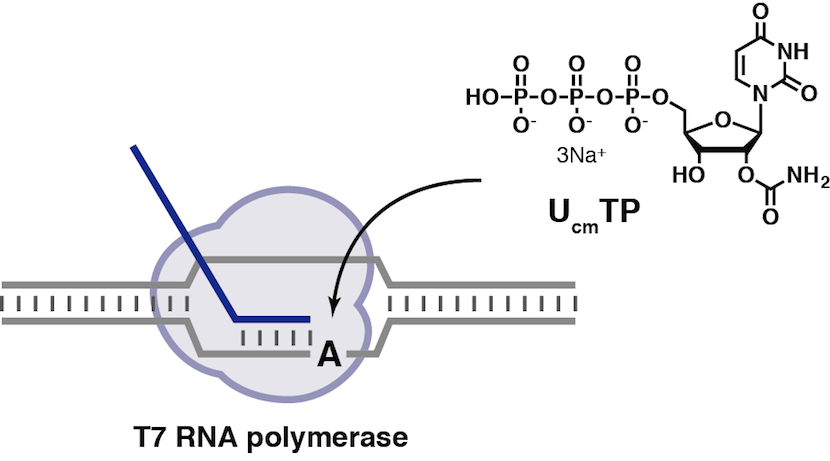
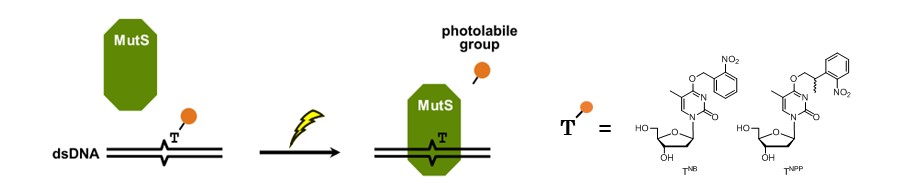
Photo-controlled binding of MutS to photo-caged DNA duplexes incorporating 4-O-(2-nitrobenzyl) or 4-O-[2-(2-nitrophenyl)propyl]thymidine
Seio K, Ohno Y, Ohno K, Takeshita L, Kanamori T, Masaki Y, Sekine M.
Bioorg Med Chem Lett. 2016 Oct 1;26(19):4861-3. doi: 10.1016/j.bmcl.2016.07.075.
An Effective Strategy for a Conformer-Selective Detection of Short-Lived Excited State Species: Application to the IR Spectroscopy of the N1H Keto Tautomer of Guanine
Asami H, Tokugawa M, Masaki Y, Ishiuchi S, Gloaguen E, Seio K, Saigusa H, Fujii M, Sekine M, Mons M.
J Phys Chem A. 2016 Apr 14;120(14):2179-84. doi: 10.1021/acs.jpca.6b01194.
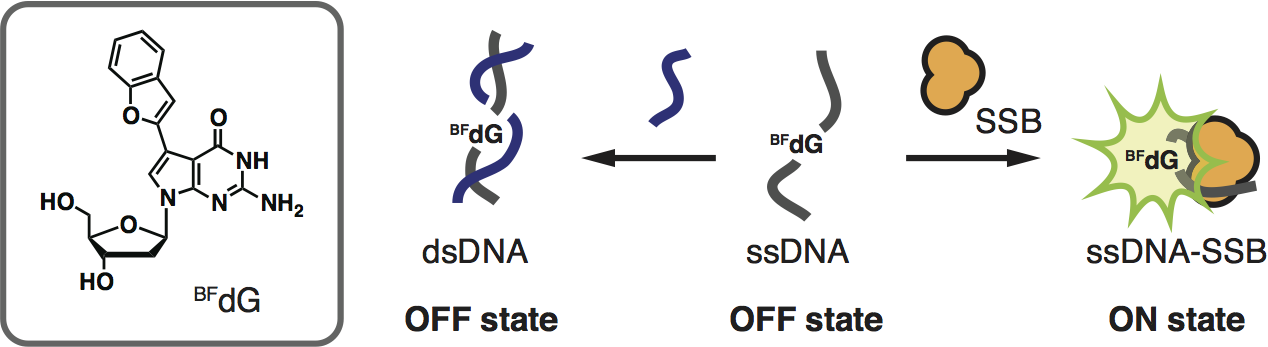
7-(Benzofuran-2-yl)-7-deazadeoxyguanosine as a fluorescence turn-ON probe for single-strand DNA binding protein
Tokugawa M, Masaki Y, Canggadibrata J C, Kaneko K, Shiozawa T, Kanamori T, Grøtli M, Wilhelmsson L M, Sekine M, Seio K.
Chem Commun (Camb). 2016 Mar 7;52(19):3809-12. doi: 10.1039/c5cc09700b.
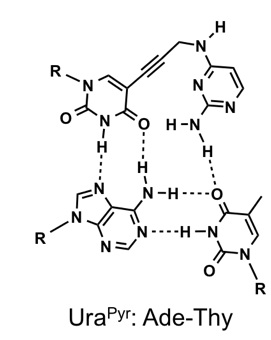
Synthesis of 5-[3-(2-aminopyrimidin-4-yl)aminopropyn-1-yl]uracil derivative that recognizes Ade-Thy base pairs in double-stranded DNA
Ito Y, Masaki Y, Kanamori T, Ohkubo A, Seio K, Sekine M.
Bioorg Med Chem Lett. 2016 Jan 1;26(1):194-6. doi: 10.1016/j.bmcl.2015.11.003.
2015
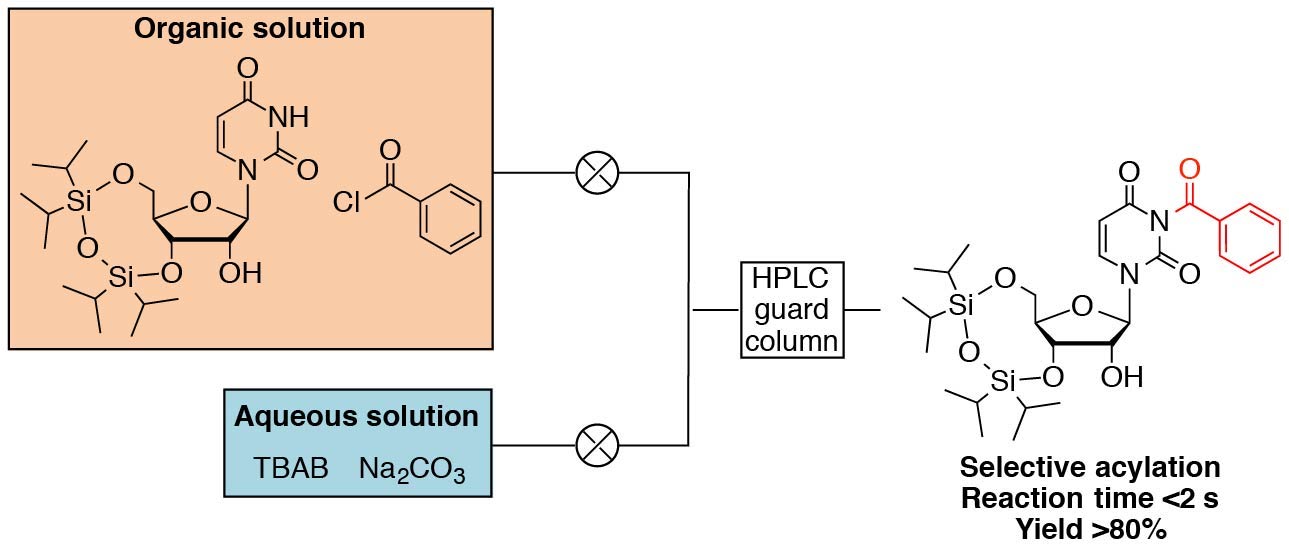
A New Microfluidic Phase-Transfer Reaction Using HPLC Guard Columns as the Reactor for the N3-Protection of Uridine Derivatives
Tago N, Masaki Y, Nagasawa H, Kanamori T, Ohkubo A, Seio K, Sekine M.
Synlett. 2015 26(18): 2578-2582. doi:10.1055/s-0035-1560264
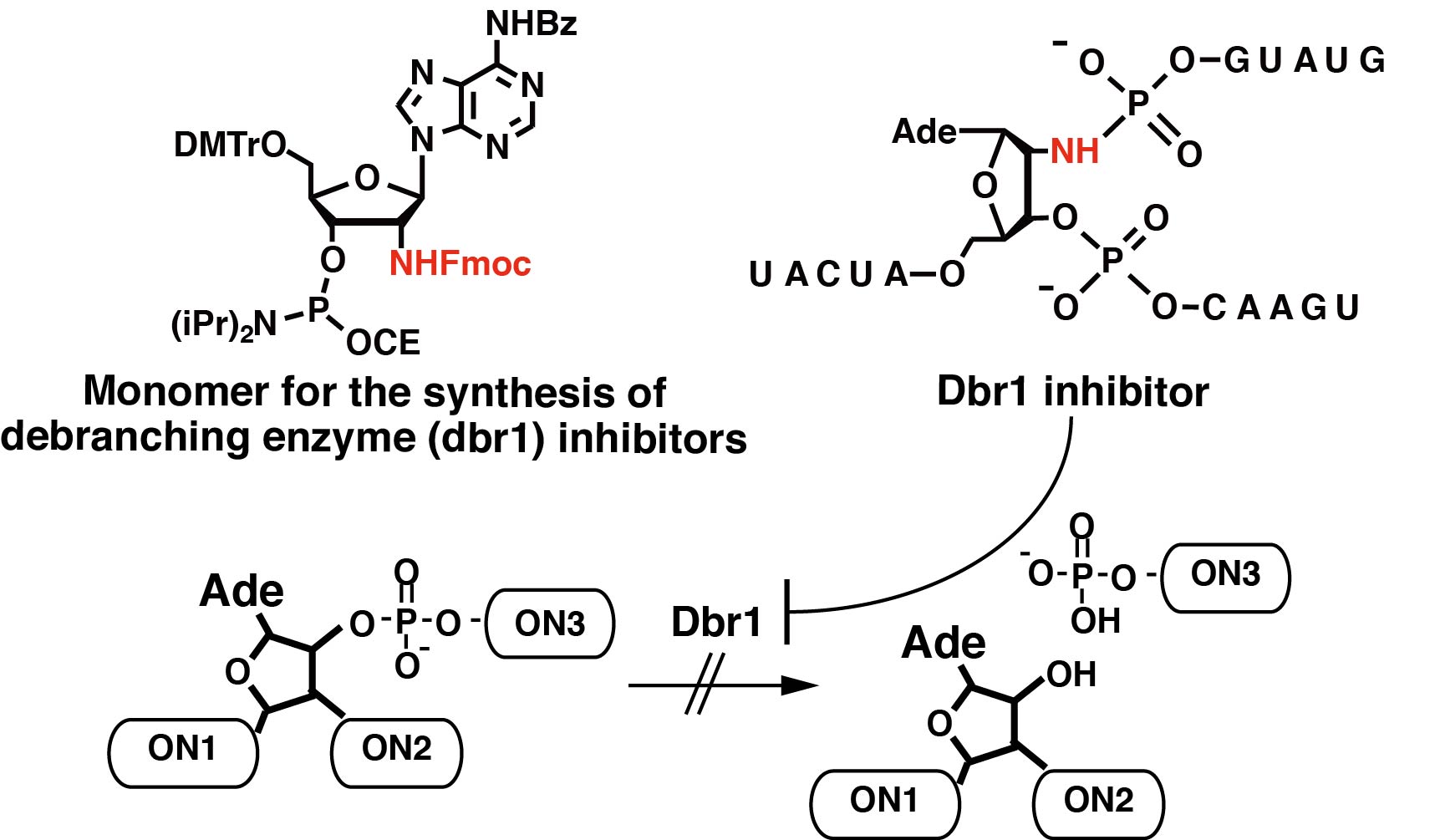
Design, Synthesis, and Properties of Phosphoramidate 2′,5′-Linked Branched RNA: Toward the Rational Design of Inhibitors of the RNA Lariat Debranching Enzyme
Tago N, Katolik A, Clark N E, Montemayor E J, Seio K, Sekine M. Hart P J, Damha M J.
J Org Chem. 2015 Oct 16;80(20):10108-18. doi: 10.1021/acs.joc.5b01719.
Synthesis and triplex-forming properties of oligonucleotides capable of recognizing corresponding DNA duplexes containing four base pairs
Ohkubo A, Yamada K, Ito Y, Yoshimura K, Miyauchi K, Kanamori T, Masaki Y, Seio K, Yuasa H, Sekine M.
Nucleic Acids Res., 2015 May 3;43:5675-5686. doi:10.1093/nar/gkv496
Synthesis of Peptide Nucleic Acids Containing Pyridazine Derivatives As Cytosine and Thymine Analogs, and Their Duplexes with Complementary Oligodeoxynucleotides
Tomori T, Miyatake Y, Sato Y, Kanamori T, Masaki Y, Ohkubo A, Sekine M, Seio K.
Org. Lett., 2015 Mar 10;17:1609–1612. doi:10.1021/acs.orglett.5b00522
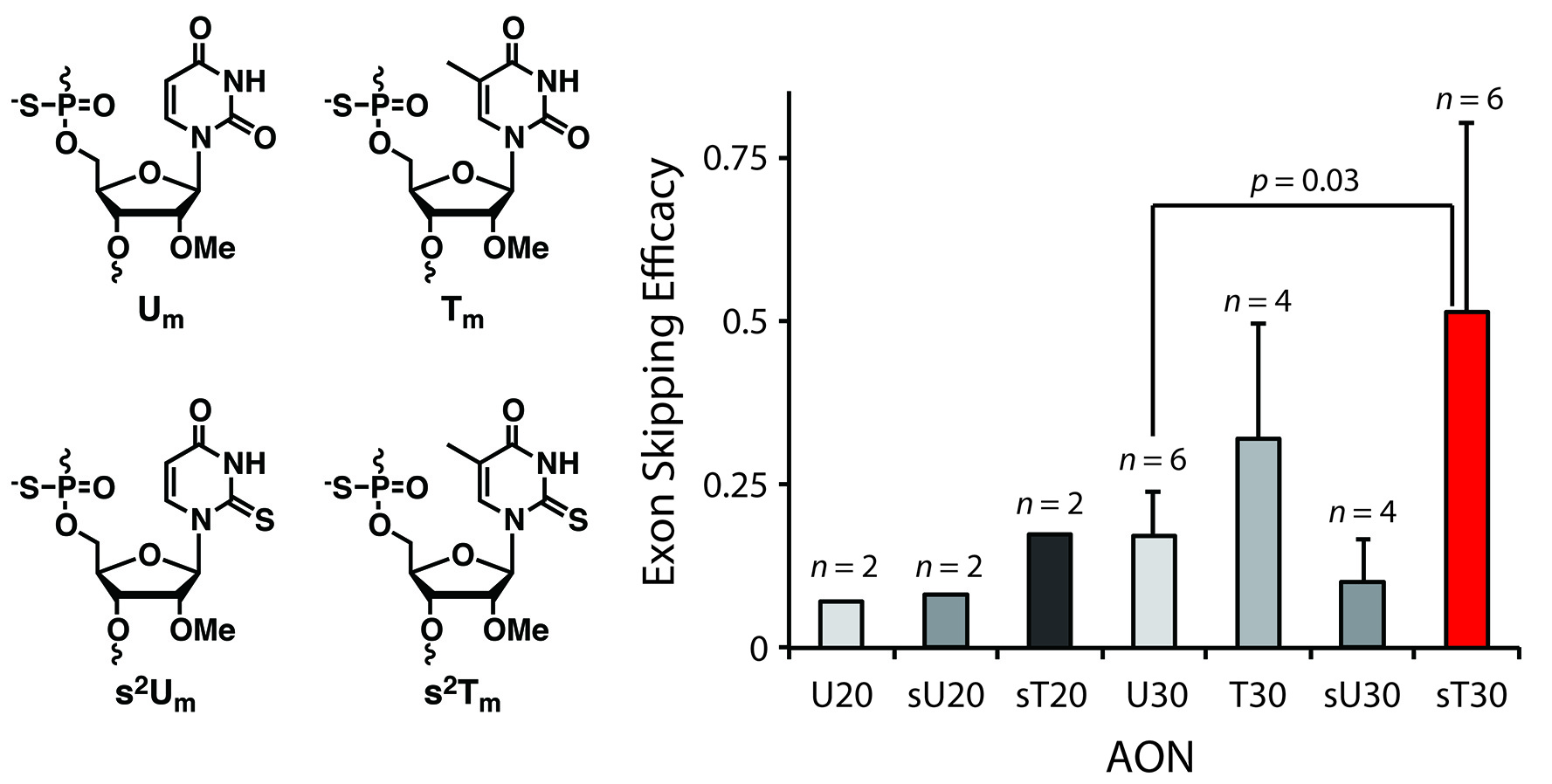
Enhancement of exon skipping in mdx52 mice by 2´-O-methyl- 2-thioribothymidine incorporation into phosphorothioate oligonucleotides
Masaki Y, Inde T, Nagata T, Tanihata J, Kanamori T, Seio K, Takeda S, Sekine M.
Med. Chem. Commun., 2015 Apr 1;6:630-633. doi:10.1039/C4MD00468J
Synthesis of Responsive Fluorescent Nucleobases 7-(Benzofuran-2-yl)-7-deazahypoxanthine and 7-(Benzofuran-2-yl)-7-deazaguanine using Cross-coupling Reaction
Tokugawa M, Kaneko K, Saito M, Kanamori T, Masaki Y, Ohkubo A, Sekine M, Seio K.
Chem Lett. 2015 Jan 5;44:64-66. doi:10.1246/cl.140879
Controlling the Fluorescence of Benzofuran-Modified Uracil Residues in Oligonucleotides by Triple-Helix Formation
Kanamori T, Ohzeki H, Masaki Y, Ohkubo A, Takahashi M, Tsuda K, Ito T, Shirouzu M, Kuwasako K, Muto Y, Sekine M, Seio K.
ChemBioChem. 2015 Jan 2;16:167-176. doi:10.1002/cbic.201402346
2014
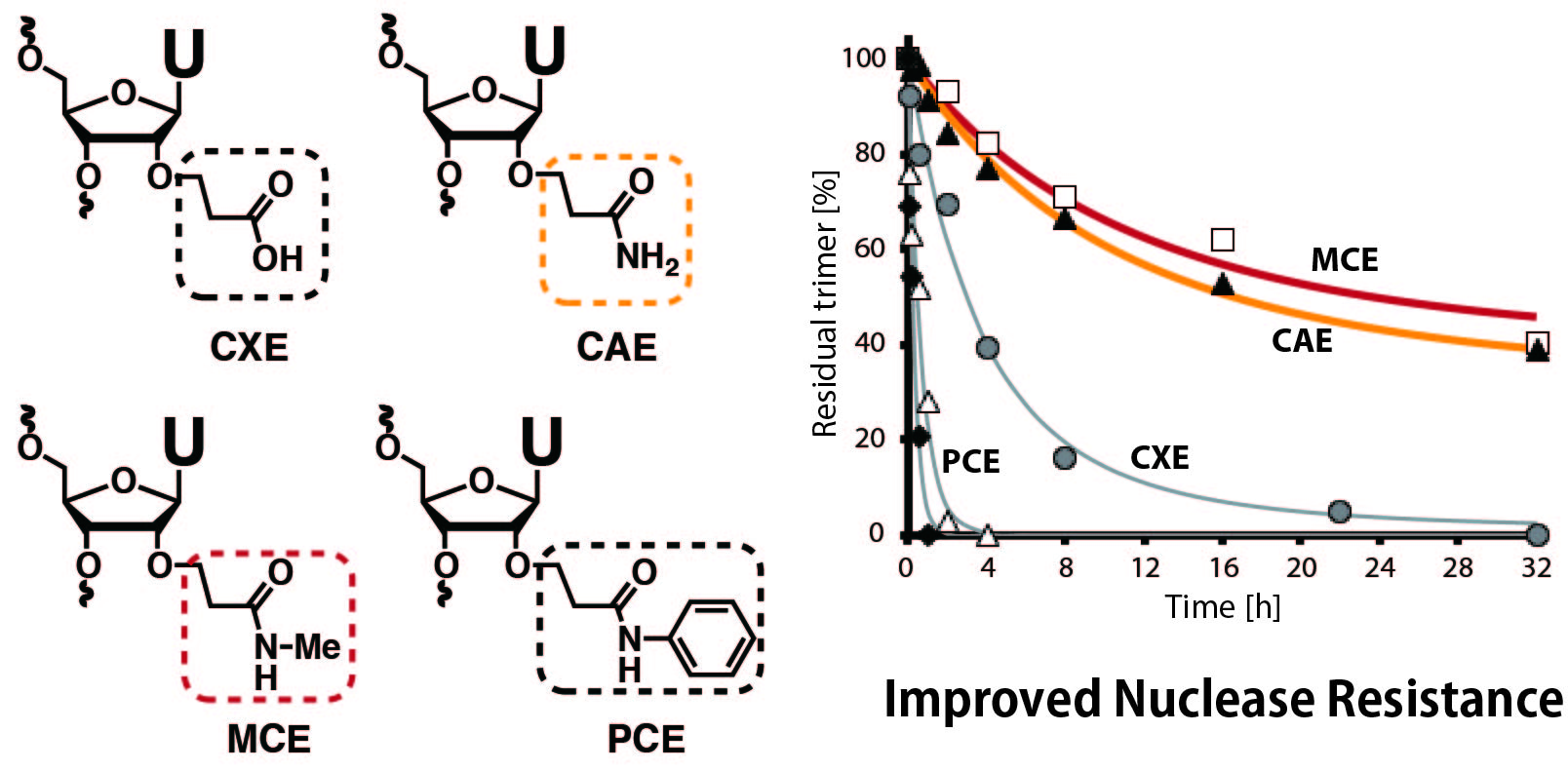
Synthesis and properties of oligonucleotides modified with 2´-O-(2-carboxyethyl)nucleotides and their carbamoyl derivatives
Yamada T, Masaki Y, Okaniwa N, Kanamori T, Ohkubo A, Tsunoda H, Seio K, Sekine M.
Org Biomol Chem. 2014 Jun 27;12(33):6457-6464 doi:10.1039/C4OB01260G
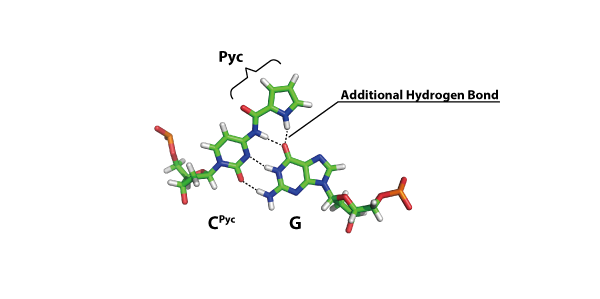
A new modified cytosine base capable of base pairing with guanine using four hydrogen bonds
Yamada K, Masaki Y, Tsunoda H, Ohkubo A, Seio K, Sekine M.
Org Biomol Chem. 2014 Feb 11;12(14):2255-2262 doi:10.1039/C3OB42420K
Properties of 5- and/or 2-modified 2´-O-cyanoethyl uridine residue : 2´-O-cyanoethyl-5-propynyl-2-thiouridine as an efficient duplex stabilizing component
Masaki Y, Miyasaka R, Hirai K, Kanamori T, Tsunoda H, Ohkubo A, Seio K, Sekine M.
Org Biomol Chem. 2014 Feb 21;12(7):1157-1162 doi:10.1039/C3OB41983E
2013
Assembly of pyrene-modified DNA/RNA duplexes incorporating a G-rich single strand region
Seio K, Tokugawa M, Tsunoda H, Ohkubo A, Arisaka H, Sekine M.
Bioorg Med Chem Lett. 2013 Dec 15;23(24):6822-6824. doi: 10.1016/j.bmcl.2013.10.012.
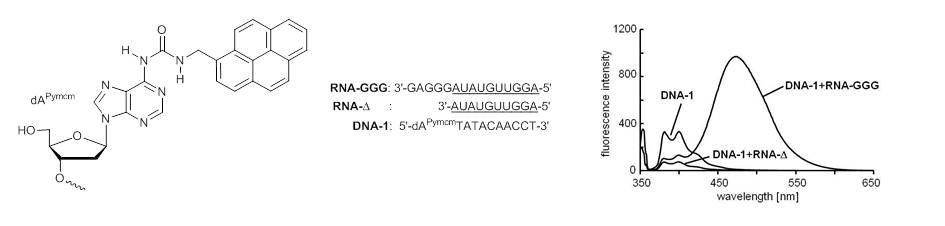
Modified oligodeoxynucleotide primers for reverse-transcription of target RNAs that can discriminate among length variants at the 3′-terminus.
Iijima Y, Kojima S, Kodama E, Kurohagi S, Kanamori T, Masaki Y, Ohkubo A, Sekine M, Seio K.
Org Biomol Chem. 2013 Oct 2;11(47):8276-8282 doi:10.1039/C3OB41901K
Chemical synthesis of U1 snRNA derivatives.
Ohkubo A, Kondo Y, Suzuki M, Kobayashi H, Kanamori T, Masaki Y, Seio K, Nagai K, Sekine M.
Org Lett. 2013 August 16;15(17):4386-4389 doi:10.1021/ol401917r
Fluorescent properties of oligonucleotides doubly modified with an indole-fused cytosine analog and 2-aminopurine.
Seio K, Kanamori T, Tokugawa M, Ohzeki H, Masaki Y, Tsunoda H, Ohkubo A, Sekine M.
Bioorg Med Chem. 2013 June 1;21(11):3197-3201. doi: 10.1016/j.bmc.2013.03.034.
Base recognition of gap sites in DNA–DNA and DNA–RNA duplexes by short oligonucleotides.
Yamada K, Ohkubo A, Esaka Y, Kanamori T, Masaki Y, Seio K, Sekine M.
Bioorg Med Chem Lett. 2013 June 1;23(11):3448-3451. doi: 10.1016/j.bmcl.2013.03.054.
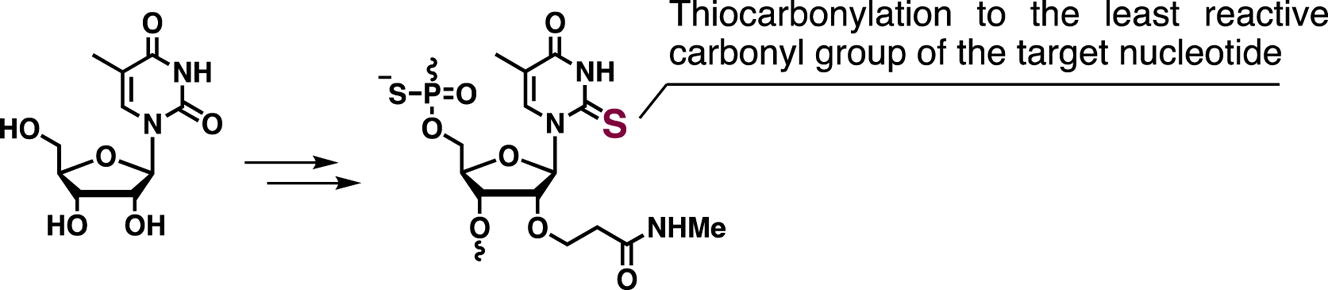
Remarkable stabilization of antiparallel DNA triplexes by strong stacking effects of consecutively modified nucleobases containing thiocarbonyl groups.
Yamada K, Hattori Y, Inde T, Kanamori T, Ohkubo A, Seio K, Sekine M.
Bioorg Med Chem Lett. 2013 Feb 1;23(3):776-778. pii: S0960-894X(12)01531-4. doi: 10.1016/j.bmcl.2012.11.079.
Topological Switch-OFF and β-Galactosidase-Triggered Switch-ON of Cyclic Antisense Oligonucleotides via CuAAC for Controlled RNA Cleavage
Miyaji K, Takeuchi K, Seio K.
Bioconjugate Chem. 2025 Aug 20; 36(8):1820-1837 doi.org/10.1021/acs.bioconjchem.5c00295.

Synthesis of Prodrug-Type Oligonucleotides Modified With a Galactosylated Self-Immolative Linker Cleavable by β-Galactosidase
Miyaji K, Masaki Y, Seio K.
Curr Protoc. 2025 Apr 7;5(4):e70128. doi: 10.1002/cpz1.70128.
Synthesis of LNA gapmers that replace a phosphorothioate linkage with a sulfonamide in the gap region, and their ability to form duplexes with complementary RNA targets
Seio K, Ohnishi R, Tachibana S, Mikagi H, Masaki Y.
Org. Biomol. Chem. 2025 Jan 2;23(2):400-409. doi.org/10.1039/d4ob01350f.
2024
Inhibitory Effects on RNA Binding and RNase H Induction Activity of Prodrug-Type Oligodeoxynucleotides Modified with a Galactosylated Self-Immolative Linker Cleavable by β-Galactosidase
Miyaji K, Masaki Y, Seio K.
Bioconjugate Chem. 2024 Oct 8; 35(10):1587-1607. doi.org/10.1021/acs.bioconjchem.4c00376.

2023
Synthesis and Conformational Analyses of Cyclonucleoside Having 13-Membered Ring Bridging Nucleobase and 5′-Position via a Linker Containing Sulfonamide
Kanagawa T, Tachibana S, Masaki Y, Seio K.
Org. Lett. 2023 Nov 3; 25:7868-7872. doi.org/10.1021/acs.orglett.3c03094.

Alteration of target cleavage patterns and off-target reduction of antisense oligonucleotides incorporating 2-N-carbamoyl- or (2-pyridyl)guanine
Kanagawa T, Koyama A, Masaki Y, Seio K.
Org. Biomol. Chem. 2023 Jun 28; 21:5214-5224. doi.org/10.1039/d3ob00574g.
2022
Insertion of a methylene group into the backbone of an antisense oligonucleotide reveals the importance of deoxyribose recognition by RNase H.
Masaki Y, Tabira A, Hattori S, Wakatsuki S, Seio K.
Org. Biomol Chem. 2022 Oct 27;20:8917-8924. doi.org/10.1039/D2OB01667B.
Quantification of synthetic errors during chemical synthesis of DNA and its suppression by non-canonical nucleosides.
Masaki Y, Onishi Y, Seio K.
Sci Rep. 2022 Jul 15;12(1):12095. doi: 10.1038/s41598-022-16222-2.
Selective and stable base pairing by alkynylated nucleosides featuring a spatially-separated recognition interface.
Okamura H, Trinh G, Dong Z, Masaki Y, Seio K, Nagatsugi F.
Nucleic Acids Res. 2022 Feb 15. doi: 10.1093/nar/gkac140
Oligodeoxynucleotides Modified with 2'- O-(Cysteinylaminobutyl)carbamoylethylribothymidine Residues for Native Chemical Ligation with Peptide at Internal Positions.
Masaki Y, Maruyama A, Yoshida K, Tomori T, Kishimura T, Seio K.
Bioconjug. Chem. 2022 Feb 7;33(2):272-278. doi: 10.1021/acs.bioconjchem.1c00575

2021
Synthesis of 2′-O-alkylcarbamoylethyl-modified oligonucleotides with enhanced nuclease resistance that form isostable duplexes with complementary RNA.
Kishimura T, Tomori T, Masaki Y, Seio K.
Bioorg Med Chem Lett. 2021 Mar 1;35:127779. doi: 10.1016/j.bmcl.2021.127779

2020
Novel EGFP reporter cell and mouse models for sensitive imaging and quantification of exon skipping
Hara Y, Mizobe Y, Inoue Y, Hashimoto Y, Motohashi N, Masaki Y, Seio K, Takeda S, Nagata T, Wood M, Inoue T, Aoki Y.
Scientific Reports 2020 Jun 22;10. doi: 10.1038/s41598-020-67077-4
Transcription of DNA duplex containing deoxypseudouridine and deoxypseudoisocytidine, and inhibition of transcription by triplex forming oligonucleotide that recognizes the modified duplex
Seio K, Yamaguchi K, Yamazaki A, Kanamori T, Masaki Y.
Nucleosides, Nucleotides & Nucleic Acids 2020 Mar 4;39(6):892-904. doi: 10.1080/15257770.2020.1714652
Synthesis of Deoxypseudouridine 5′-Triphosphate Bearing the Photoremovable Protecting Group at the N1 Position Capable of Enzymatic Incorporation to DNA
Takeshita L, Yamada Y, Masaki Y, Seio K.
J. Org. Chem. 2020 Jan 7;85:1861-1870. doi: 10.1021/acs.joc.9b02194

2019
Modification of oligonucleotides with weak basic residues via the 2′-O-carbamoylethyl linker for improving nuclease resistance without loss of duplex stability and antisense activity
Masaki Y, Yamamoto K, Yoshida K, Maruyama A, Tomori T, Iriyama Y, Nakajima H, Kanaki T, Seio K.
Org. Biomol Chem. 2019 Apr 29;17:4835-4842. doi: 10.1039/c9ob00668k

31P NMR Study on the Reactions of Amino Acids and Sugar Derivatives with Pyrophosphorous Acid as a Possible Prebiotic Phosphonylating Agent
Seio K, Shiozawa T, Sugiyama D, Ohno K, Tomori T, Masaki Y.
Bull. Chem. Soc. Jpn. 2019 Apr 10;92(4):905-911. doi: 10.1246/bcsj.20180392

DNA triplex-based fluorescence turn-on sensors for adenosine using a fluorescent molecular rotor 5-(3-methylbenzofuran-2-yl) deoxyuridine
Kanamori T, Masaki Y, Oda Y, Ohzeki H, Ohkubo A, Sekine M, Seio K.
Org. Biomol Chem. 2019 Jan 25;17:2077-2080. doi: 10.1039/C8OB02747A

Synthesis of 2′-O-(N-methylcarbamoylethyl) 5-methyl-2-thiouridine and its application to splice-switching oligonucleotides.
Masaki Y, Yamamoto K, Inde T, Yoshida K, Maruyama A, Nagata T, Tanihata J, Takeda S, Sekine M, Seio K.
Bioorg Med Chem Lett. 2019 Jan 15;29(2):160-163. doi: 10.1016/j.bmcl.2018.12.005

Tolerance of N2-heteroaryl modifications on guanine bases in a DNA G-quadruplex
Masaki Y, Inde T, Maruyama A, Seio K.
Org. Biomol Chem. 2019 Jan 4;17:859-866. doi: 10.1039/C8OB03100B

A theoretical study on the elimination reaction of acrylonitrile from 2′-O-cyanoethylated nucleosides by Bu4NF.
Ando K, Saneyoshi H, Seio K, Sekine M.
Tetrahedron 2019 Jan 3;75(1):1-9. doi: 10.1016/j.tet.2018.11.042

2018
Synthesis of and triplex formation in oligonucleotides containing 2'-deoxy-6-thioxanthosine.
Inde T, Nishizawa S, Hattori Y, Kanamori T, Yuasa H, Seio K, Sekine M, Ohkubo A.
Bioorg Med Chem. 2018 Jul 30;26(13):3785-3790. doi: 10.1016/j.bmc.2018.06.004

Application of 2'-O-(2-N-Methylcarbamoylethyl) Nucleotides in RNase H-Dependent Antisense Oligonucleotides.
Masaki Y, Iriyama Y, Nakajima H, Kuroda Y, Kanaki T, Furukawa S, Sekine M, Seio K.
Nucleic Acid Ther. 2018 Jul 18;28(5):307-311. doi: 10.1089/nat.2018.0738

Deoxynucleoside Triphosphate Containing Pyridazin-3-one Aglycon as a Thymidine Triphosphate Substitute for Primer Extension and Chain Elongation by Klenow fragments.
Tomori T, Nagaoka K, Takeshita L, Shiozawa T, Miyatake Y, Masaki Y, Sekine M, Seio K.
J Org Chem. 2018 Jun 28;83(15):8353-8363. doi: 10.1021/acs.joc.8b00918

Solvent- and environment-dependent fluorescence of modified nucleobases
Seio K, Kanamori T, Masaki Y.
Tetrahedron Lett. 2018 May 23;59(21):1977-1985. doi: 10.1016/j.tetlet.2018.04.003

2017
Synthesis of photocaged 6-O-(2-nitrobenzyl)guanosine and 4-O-(2-nitrobenzyl) uridine triphosphates for photocontrol of the RNA transcription reaction.
Ohno K, Sugiyama D, Takeshita L, Kanamori T, Masaki Y, Sekine M, Seio K.
Bioorg Med Chem. 2017 Sep 21. pii: S0968-0896(17)31538-9. doi: 10.1016/j.bmc.2017.09.032.

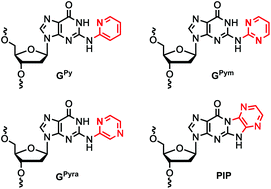
Synthesis of oligonucleotides containing 2-N-heteroarylguanine residues and their effect on duplex/triplex stability.
Inde T, Masaki Y, Maruyama A, Ito Y, Makio N, Miyatake Y, Tomori T, Sekine M, Seio K.
Org Biomol Chem. 2017 Oct 11;15(39):8371-8383. doi: 10.1039/c7ob01875d.

A Systematic Study of the Synthesis of 2ʹ-Deoxynucleosides by Mitsunobu Reaction.
Seio K, Tokugawa M, Kaneko K, Shiozawa T, Masaki, Y.
Synlett 2017; 28(15): 2014-2017. doi: 10.1055/s-0036-1588445.

Deformability Calculation for Estimation of the Relative Stability of Chemically Modified RNA Duplexes.
Masaki Y, Sekine M, Seio K.
Curr Protoc Nucleic Acid Chem. 2017 Mar 2;68:7.27.1-7.27.10.
doi:10.1002/cpnc.25.
Fluorescence enhancement of oligodeoxynucleotides modified with green fluorescent protein chromophore mimics upon triplex formation.
Kanamori T, Takamura A, Tago N, Masaki Y, Ohkubo A, Sekine M, Seio K.
Org Biomol Chem. 2017 Feb 1;15(5):1190-1197. doi:10.1039/c6ob01278g.

2016
Enzymatic synthesis and reverse transcription of RNAs incorporating 2′-O-carbamoyl uridine triphosphate
Masaki Y, Ito H, Oda Y, Yamazaki K, Tago N, Ohno K, Ishii N, Tsunoda H, Kanamori T, Ohkubo A, Sekine M, Seio K.
Chem Commun (Camb). 2016 Oct 25;52(87):12889-12892. doi:10.1039/C6CC05796A

Photo-controlled binding of MutS to photo-caged DNA duplexes incorporating 4-O-(2-nitrobenzyl) or 4-O-[2-(2-nitrophenyl)propyl]thymidine
Seio K, Ohno Y, Ohno K, Takeshita L, Kanamori T, Masaki Y, Sekine M.
Bioorg Med Chem Lett. 2016 Oct 1;26(19):4861-3. doi: 10.1016/j.bmcl.2016.07.075.

An Effective Strategy for a Conformer-Selective Detection of Short-Lived Excited State Species: Application to the IR Spectroscopy of the N1H Keto Tautomer of Guanine
Asami H, Tokugawa M, Masaki Y, Ishiuchi S, Gloaguen E, Seio K, Saigusa H, Fujii M, Sekine M, Mons M.
J Phys Chem A. 2016 Apr 14;120(14):2179-84. doi: 10.1021/acs.jpca.6b01194.
7-(Benzofuran-2-yl)-7-deazadeoxyguanosine as a fluorescence turn-ON probe for single-strand DNA binding protein
Tokugawa M, Masaki Y, Canggadibrata J C, Kaneko K, Shiozawa T, Kanamori T, Grøtli M, Wilhelmsson L M, Sekine M, Seio K.
Chem Commun (Camb). 2016 Mar 7;52(19):3809-12. doi: 10.1039/c5cc09700b.

Synthesis of 5-[3-(2-aminopyrimidin-4-yl)aminopropyn-1-yl]uracil derivative that recognizes Ade-Thy base pairs in double-stranded DNA
Ito Y, Masaki Y, Kanamori T, Ohkubo A, Seio K, Sekine M.
Bioorg Med Chem Lett. 2016 Jan 1;26(1):194-6. doi: 10.1016/j.bmcl.2015.11.003.

2015
A New Microfluidic Phase-Transfer Reaction Using HPLC Guard Columns as the Reactor for the N3-Protection of Uridine Derivatives
Tago N, Masaki Y, Nagasawa H, Kanamori T, Ohkubo A, Seio K, Sekine M.
Synlett. 2015 26(18): 2578-2582. doi:10.1055/s-0035-1560264

Design, Synthesis, and Properties of Phosphoramidate 2′,5′-Linked Branched RNA: Toward the Rational Design of Inhibitors of the RNA Lariat Debranching Enzyme
Tago N, Katolik A, Clark N E, Montemayor E J, Seio K, Sekine M. Hart P J, Damha M J.
J Org Chem. 2015 Oct 16;80(20):10108-18. doi: 10.1021/acs.joc.5b01719.

Synthesis and triplex-forming properties of oligonucleotides capable of recognizing corresponding DNA duplexes containing four base pairs
Ohkubo A, Yamada K, Ito Y, Yoshimura K, Miyauchi K, Kanamori T, Masaki Y, Seio K, Yuasa H, Sekine M.
Nucleic Acids Res., 2015 May 3;43:5675-5686. doi:10.1093/nar/gkv496
Synthesis of Peptide Nucleic Acids Containing Pyridazine Derivatives As Cytosine and Thymine Analogs, and Their Duplexes with Complementary Oligodeoxynucleotides
Tomori T, Miyatake Y, Sato Y, Kanamori T, Masaki Y, Ohkubo A, Sekine M, Seio K.
Org. Lett., 2015 Mar 10;17:1609–1612. doi:10.1021/acs.orglett.5b00522
Enhancement of exon skipping in mdx52 mice by 2´-O-methyl- 2-thioribothymidine incorporation into phosphorothioate oligonucleotides
Masaki Y, Inde T, Nagata T, Tanihata J, Kanamori T, Seio K, Takeda S, Sekine M.
Med. Chem. Commun., 2015 Apr 1;6:630-633. doi:10.1039/C4MD00468J

Synthesis of Responsive Fluorescent Nucleobases 7-(Benzofuran-2-yl)-7-deazahypoxanthine and 7-(Benzofuran-2-yl)-7-deazaguanine using Cross-coupling Reaction
Tokugawa M, Kaneko K, Saito M, Kanamori T, Masaki Y, Ohkubo A, Sekine M, Seio K.
Chem Lett. 2015 Jan 5;44:64-66. doi:10.1246/cl.140879

Controlling the Fluorescence of Benzofuran-Modified Uracil Residues in Oligonucleotides by Triple-Helix Formation
Kanamori T, Ohzeki H, Masaki Y, Ohkubo A, Takahashi M, Tsuda K, Ito T, Shirouzu M, Kuwasako K, Muto Y, Sekine M, Seio K.
ChemBioChem. 2015 Jan 2;16:167-176. doi:10.1002/cbic.201402346
2014
Synthesis and properties of oligonucleotides modified with 2´-O-(2-carboxyethyl)nucleotides and their carbamoyl derivatives
Yamada T, Masaki Y, Okaniwa N, Kanamori T, Ohkubo A, Tsunoda H, Seio K, Sekine M.
Org Biomol Chem. 2014 Jun 27;12(33):6457-6464 doi:10.1039/C4OB01260G

A new modified cytosine base capable of base pairing with guanine using four hydrogen bonds
Yamada K, Masaki Y, Tsunoda H, Ohkubo A, Seio K, Sekine M.
Org Biomol Chem. 2014 Feb 11;12(14):2255-2262 doi:10.1039/C3OB42420K

Properties of 5- and/or 2-modified 2´-O-cyanoethyl uridine residue : 2´-O-cyanoethyl-5-propynyl-2-thiouridine as an efficient duplex stabilizing component
Masaki Y, Miyasaka R, Hirai K, Kanamori T, Tsunoda H, Ohkubo A, Seio K, Sekine M.
Org Biomol Chem. 2014 Feb 21;12(7):1157-1162 doi:10.1039/C3OB41983E

2013
Assembly of pyrene-modified DNA/RNA duplexes incorporating a G-rich single strand region
Seio K, Tokugawa M, Tsunoda H, Ohkubo A, Arisaka H, Sekine M.
Bioorg Med Chem Lett. 2013 Dec 15;23(24):6822-6824. doi: 10.1016/j.bmcl.2013.10.012.
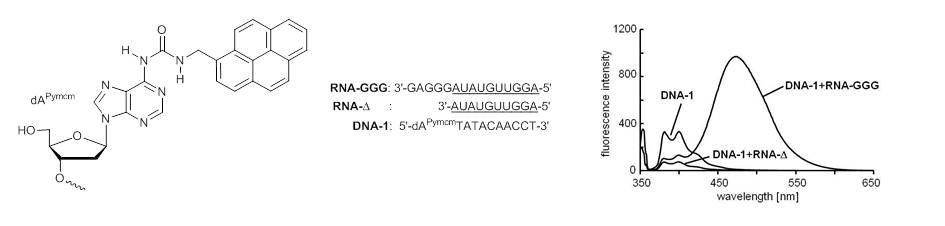
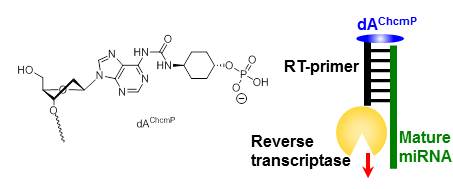
Modified oligodeoxynucleotide primers for reverse-transcription of target RNAs that can discriminate among length variants at the 3′-terminus.
Iijima Y, Kojima S, Kodama E, Kurohagi S, Kanamori T, Masaki Y, Ohkubo A, Sekine M, Seio K.
Org Biomol Chem. 2013 Oct 2;11(47):8276-8282 doi:10.1039/C3OB41901K

Chemical synthesis of U1 snRNA derivatives.
Ohkubo A, Kondo Y, Suzuki M, Kobayashi H, Kanamori T, Masaki Y, Seio K, Nagai K, Sekine M.
Org Lett. 2013 August 16;15(17):4386-4389 doi:10.1021/ol401917r

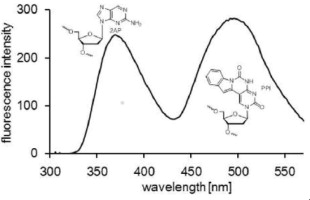
Fluorescent properties of oligonucleotides doubly modified with an indole-fused cytosine analog and 2-aminopurine.
Seio K, Kanamori T, Tokugawa M, Ohzeki H, Masaki Y, Tsunoda H, Ohkubo A, Sekine M.
Bioorg Med Chem. 2013 June 1;21(11):3197-3201. doi: 10.1016/j.bmc.2013.03.034.

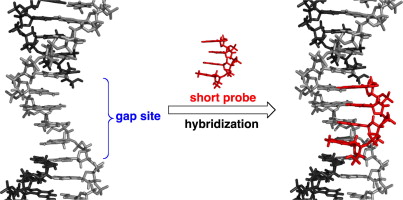
Base recognition of gap sites in DNA–DNA and DNA–RNA duplexes by short oligonucleotides.
Yamada K, Ohkubo A, Esaka Y, Kanamori T, Masaki Y, Seio K, Sekine M.
Bioorg Med Chem Lett. 2013 June 1;23(11):3448-3451. doi: 10.1016/j.bmcl.2013.03.054.

Remarkable stabilization of antiparallel DNA triplexes by strong stacking effects of consecutively modified nucleobases containing thiocarbonyl groups.
Yamada K, Hattori Y, Inde T, Kanamori T, Ohkubo A, Seio K, Sekine M.
Bioorg Med Chem Lett. 2013 Feb 1;23(3):776-778. pii: S0960-894X(12)01531-4. doi: 10.1016/j.bmcl.2012.11.079.
